Brains make sense and perceptions from the signals that flow through them. Can we identify elementary neural “wiring” configurations and interactions that help us to understand how brains accomplish this feat?


Cortical tissue is packed tight with neurons, dendrites, and axons as illustrated in the electron microscope image of a slice of mouse neocortex in Figure 1 above. To better comprehend this tangle, neuroscientists have worked to identify elementary categories of interactions between neurons and the ways those neurons are wired together.
Neurons in cortical tissue can be divided into excitatory and inhibitory cells as in Figure 2 at right (red for excitatory and blue for inhibitory). This schematic diagram shows interactions in the context of elementary connection motifs. The figure is from a recent paper summarizing evidence the mammalian neocortex is an elaboration of a brain area from a common ancestor of mammals, reptiles, and birds thought to have existed about 320 million years ago (“Neocortical Lamination: Insights from Neuron Types and Evolutionary Precursors” published November 7, 2017 in Frontiers in Neuroanatomy).
Research on the evolutionary origin of or our six-layered neocortex may provide us glimpses into why specific neuron types or connection motifs provided selective advantages to our ancestors. Evidence from comparative anatomy and physiology suggests that the common ancestor had a three-layered brain region which was lost in birds, remained three-layered in reptiles, and developed into six-layered neocortex in mammals. Layered brain areas in today’s reptiles (turtles) and mammals (mice) that are thought to have a common origin are highlighted in blue in Figure 3 below.
Figure 2. A schematic diagram of currently established elementary circuit configurations. The red triangle represents the cell body of a pyramidal cell. The thick trunk reaching up and ending with two thinner branches reaching diagonally is the pyramidal cell’s apical dendrite. The branches reaching diagonally down from the triangle’s two lower corners represent its basal dendrites. An axon leaves the bottom center of the pyramidal cell to various destinations (see text). Two other axons (red) enter the diagram. The axon that enters from right comes from other cortical areas. The axon that enters at left from below carries sensory input from the visual thalamus. Two inhibitory interneurons (blue) complete the circuit. ffexc: feed forward excitation, fbexc: feed back excitation, lexc: lateral excitation, ffinh: feed forward inhibition, fbinh: feedback inhibition. Figure 2(A) from “Neocortical Lamination: Insights from Neuron Types and Evolutionary Precursors” published November 7, 2017 in Frontiers in Neuroanatomy.
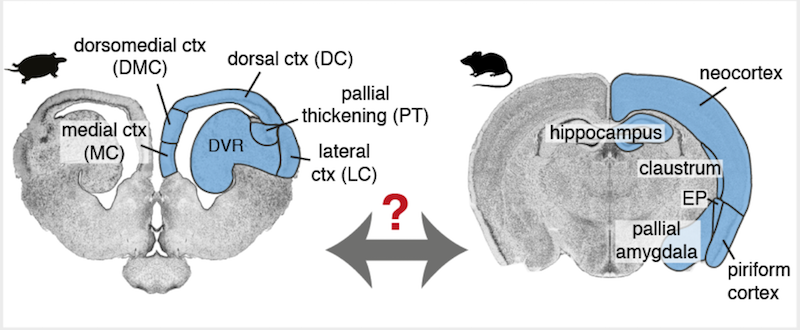
Single-cell RNA sequencing was used to investigate the evolution of brain regions thought to have common origins (blue in Figure 3) in a study published in Science(“Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles” published May 25, 2018). Genetic techniques provide some of the most direct methods available to investigate ancestral relationships among brain tissues. First, scientists pulled sequences of expressed genes from identified neuron types residing in specific regions of turtle and lizard brains. Next they looked in mammalian brains to see where those same gene sequences were expressed and in which neuron types.

Researchers found inhibitory neurons express similar sets of genes in reptiles and mammals, indicating a common ancestry. In contrast, excitatory neurons differ substantially in gene expression across these two groups. Their results are consistent with Shepherd and Rowe’s idea that three-layer cortex was co-opted and transformed into six-layer cortex through the emergence of new types of excitatory neurons after the ancestral excitatory neurons went through profound changes of gene expression. The basic circuit module seen in the ancestral three-layer cortex in Figure 2 is transformed in the mammalian neocortex into layered versions of the basic circuit with three distinct genetically expressed versions of excitatory pyramidal cells as shown in Figure 4.
In summary, recent research has provided a putative common ancestral circuit, the basic cortical module, and three variants in mammalian neocortex. Studies that compare and contrast signal processing in these four circuits could provide insights into how brains make sense and perceptions from the signals that flow through them.



