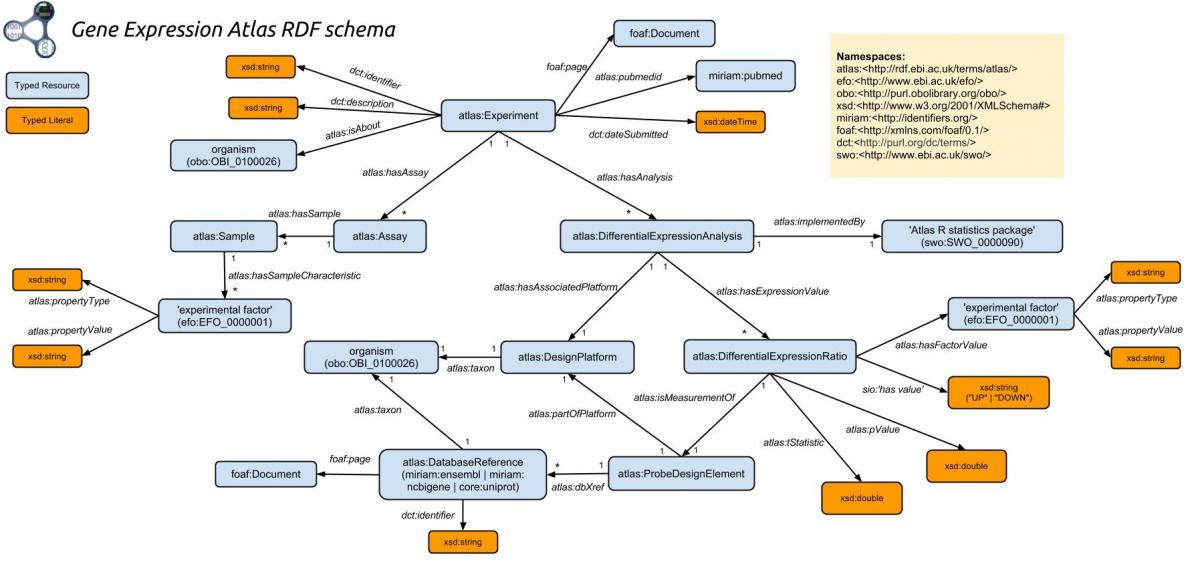
Figure 1. The Gene Expression Atlas RDF Schema. Image courtesy of EMBL-EBI.
A key idea behind the Semantic Web of linked-data is to enable seamless use of data from diverse sources across the Internet. Biological pathways data come from a huge number of laboratories from around the world and are captured and saved in a lot of different ways. We can use SPARQL endpoints to bring data together from any number of data repositories.
In this post we’ll use the R package SPARQL to pull biological pathway data from WikiPathways while at the same time pulling the genes expressed in these pathways from the Gene Expression Atlas.
You may find these related posts helpful: “WikiPathways: Open Biological Pathways Data on the Semantic Web” from June 27, 2017 and “Update on R and Semantic Web Technologies” from June 30, 2017.
The Gene Expression Atlas provides data on what genes or proteins are expressed in a particular species under specific conditions. It also provides data on differential expression, the increase or decrease of gene expression or protein production under specific conditions.
Let’s look for human biological pathways that show different gene activity under Alzheimer’s disease than under normal circumstances. Differences are indicated by significant increases or decreases in gene expression.
First, look up the identifier for Alzheimer’s disease using
the EMBL-EBI Ontology Lookup Service.
Type Alzheimer’s disease into the Search EFO search box in the upper right area of the EMBL-EBI Ontology Lookup Service page (see Figure 2 above).
EFO stands for Experimental Factor Ontology.
A large list of factors appear that are associated with Alzheimer’s disease but the one we’re interested in, listed as Alzheimer’s disease, should be on the first page and provide the identifier EFO:0000249.
The following discussion assumes that you’ve installed and loaded the R package SPARQL. If not, please see the June 30, 2017 post “Update on R and Semantic Web Technologies.”
Assign the WikiPathways SPARQL endpoint URL to the endpoint variable in your R environment.
endpoint <- 'http://sparql.wikipathways.org'
Next, assign a SPARQL query to the query variable like in the following code snippet.
query <- 'PREFIX identifiers:<http://identifiers.org/ensembl/>
PREFIX atlas: <http://rdf.ebi.ac.uk/resource/atlas/>
PREFIX atlasterms: <http://rdf.ebi.ac.uk/terms/atlas/>
PREFIX efo: <http://www.ebi.ac.uk/efo/>
SELECT DISTINCT ?wpURL ?pwTitle ?expressionValue ?pvalue where {
SERVICE <https://www.ebi.ac.uk/rdf/services/atlas/sparql> {
?factor rdf:type efo:EFO_0000249 .
?value atlasterms:hasFactorValue ?factor .
?value atlasterms:isMeasurementOf ?probe .
?value atlasterms:pValue ?pvalue .
?value rdfs:label ?expressionValue .
?probe atlasterms:dbXref ?dbXref .
}
?pwElement dcterms:isPartOf ?pathway .
?pathway dc:title ?pwTitle .
?pathway dc:identifier ?wpURL .
?pwElement wp:bdbEnsembl ?dbXref .
}
ORDER BY ASC(?pvalue)'
The full statement above is sent to the WikiPathways SPARQL endpoint (the URL assigned to the endpoint variable). However, the search terms in the embedded SERVICE statement are forwarded to the Gene Expression Atlas SPAQL endpoint (the URL following SERVICE).
The first triple inside the SERVICE statement sets the ?factor variable to the Alzheimer’s disease identifier you found above. Notice that the colon was swapped out for an underscore so that EFO:0000249 became EFO_0000249.
The second through fifth triples will all have the same subject, which is assigned to the ?value variable. The first three use predicates from the atlasterms namespace. Look at the lower right quadrant of the Gene Expression Atlas RDF Schema (see Figure 1 above). Each of the three Gene Expression Atlas specific predicates (hasFactorValue, isMeasurementOf, and pValue) has atlas:DifferentialExpressionRatio as their subject.
The final triple also uses a predicate from the atlasterms namespace. dbXref is a predicate of atlas:ProbeDesignElement that is itself the object pointed to by isMeasurementOf from atlas:DifferentialExpressionRatio. The dbXref predicate provides a bridge across various databases.
Finally, enter the SPARQL function to carry out our query and assign our results to the data variable.
data <- SPARQL(endpoint, query)
Run the R summary function on the data assigned to the data variable to see how data were returned by SPARQL.
summary(data)
SPARQL returned an R data frame and assigned it to data$results.
Take a peek at the first six rows of the results by running the R head function on the data frame assigned to data$results.
head(data$results)

Figure 3. Using R and the R package SPARQL to federate data from WikiPathays and the Gene Expression Atlas
Refer to Figure 3 above to see all interactions with the R console for this exercise. As you go through analysis, you’ll find that all but 2 of the 84 pathways show decreased gene expression in Alzheimer’s disease. Two pathways show increased COL27A1 gene expression.
Mining the world’s linked-data is relatively easy using R and the R package SPARQL. You must be proficient in SPARQL, be able to navigate ontologies, and know where the SPARQL endpoints are. You did all of this to pull pathway and differential gene expression data associated with Alzheimer’s disease and used them as a single integrated dataset!